
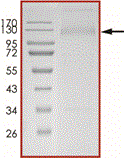
2019-nCoV Spike protein S1 (T95I, Y144T, Y145S, ins146N, R346K, E484K, N501Y, D614G, P681H)
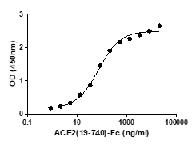
Recombinant 2019-nCoV Spike protein S1 (T95I, Y144T, Y145S, ins146N, R346K, E484K, N501Y, D614G, P681H) was expressed in CHO cells using a C-terminal His- tag.
Catalog No. C19S1-G231MH
| Catalog No. | Pack Size | Price (USD) | |
|---|---|---|---|
| C19S1-G231MH-10 | 10 ug | $100 | |
| C19S1-G231MH-20 | 20 ug | $190 | |
| C19S1-G231MH-50 | 50 ug | $290 | |
| C19S1-G231MH-100 | 100 ug | $435 | |
| C19S1-G231MH-BULK | BULK | Contact Us |

 Toll Free: 1-866-954-6273
Toll Free: 1-866-954-6273 info@signalchem.com
info@signalchem.com